New Delhi: Scientists at the Indian Institute of Science Education and Research (IISER) Kolkata have decoded the various dynamic structures of the SARS-CoV-2 spike protein — the molecular machine that allows the entry of coronavirus into our cells. The research, the scientists say, can serve as a ‘recipe’ for Covid-19 vaccine developers.
The majority of the vaccines being developed worldwide work on a simple basic principle: Exposing the body to the spike protein — which make up the protrusions seen on the outer surface of the novel coronavirus — to trick the body into believing that it has been attacked by a pathogen and hence trigger an immune response.
For this reason, the 3D structure of the spike protein was one of the first to be fully studied and characterised, and even now, researchers are trying to learn as much as possible about the spike proteins of SARS-CoV-2.
Also read: Scientists claim coronavirus breakthrough — ‘spike’ protein map could pave way for vaccine
IISER’s breakthrough
The stability of every biomolecular structure is governed by a parameter called ‘free energy’ which is fundamental in understanding how the molecule will react with its surroundings.
What the scientists at IISER Kolkata have done is to describe this free energy profile for different structures of the SARS-CoV-2 spike protein.
Susmita Roy, assistant professor of IISER Kolkata, told ThePrint that the spike proteins are made of three intertwined chains of amino acids, the building blocks of proteins.
The interactions between these chains cause a very unique set of dynamics within the spike proteins, which Roy describes as the “dance of the spike protein”. These interactions can cause the top ends of each chain of amino acid to unfurl into an ‘up’ position or fold into a ‘down’ position. The continuous shift from up to down positions make up the ‘dance’.
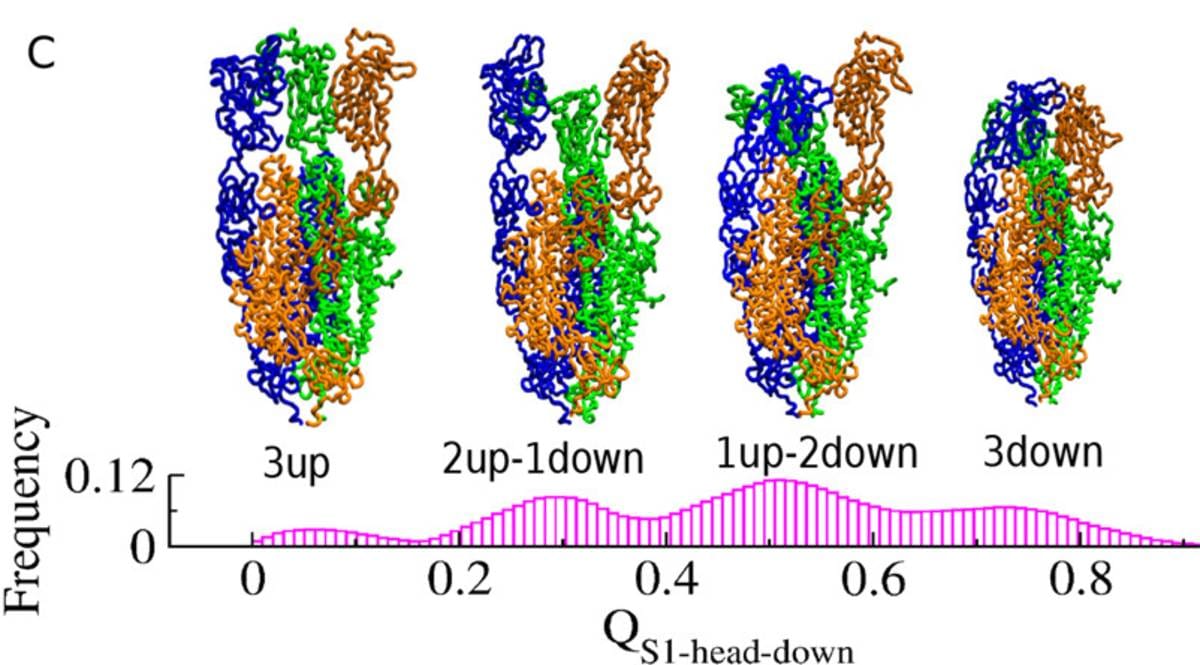
In a study published in The Journal of Physical Chemistry Letters, the team identified the states in which spike proteins are most likely to help the SARS-CoV-2 hook on to host human cells.
“When the chains are upwards, they are more exposed, which helps them to find and bind to the host cell receptors,” Roy said.
According to the researchers, these states trigger the human body’s immune response, and so understanding it can help design vaccines that induce a better immune response.
The team used existing data on the structure of the spike protein to find the ‘free energy’ associated with the different structures and determined which structures lead to a higher affinity towards the ACE2 receptors, the enzyme with which the spike protein binds to gain entry into human cells.
“In our study, we have found all the possible structural ensembles in spike proteins. We found that when either one or two chains are up, then the ACE2 receptors can bind very well,” Roy said.
Also read: Unique protein in placenta could hold key to protecting organs from Covid infection
How the study can help
The interaction-level information of the spike protein could provide deep insight into developing effective therapeutic targets as well as design vaccines, the researchers said in the study.
Most of the vaccine designs mimic the coronavirus spike protein, tricking the body into believing that it is under attack by a virus. As a result the body launches an immune response, and also learns to recognise the spike protein. This allows the body to be prepared with a more robust immune response the next time it encounters the real, live virus.
“We can say that our study is like a recipe for manipulating the spike proteins. Now that we know the atomic level mechanism of the spike protein, vaccine manufacturers can tweak the amino acid chains to get the best configuration of the spike protein,” Roy said.
This is not the first time a team has attempted to decode the molecular level structure of the spike protein. But, these earlier studies used Cryogenic electron microscopy (Cryo-EM) which failed to capture the changing dynamics of the structures.
The IISER Kolkata team’s computational study, though, was able to create a detailed map of the structures’ change in response to interactions within the spike protein.
The study also highlights some of the differences between the SARS-CoV-2 spike structures and the spike protein of the SARS virus that caused a pandemic in 2003.
“Although the structures are very similar, the dynamics of the spike proteins are different,” Roy said.
The up-down movement in the SARS-CoV-2 virus is more frequent compared to the dynamics of the spike protein in SARS and MERS viruses, which may be one of the reasons why the novel coronavirus spread as fast as it did, she added.
Also read: The world’s most powerful supercomputers have joined race to stop coronavirus






